Table Of Content
This option is useful if you want a primer to a span specific junction on the template. Note that this option cannot be used in association with the "Exon/intron selection" options above. A higher E value should be used if you want more stringent specificity checking (i.e., to identify targets that have more mismatches to the primers, in addition to the perfectly matched targets).
Best Primer design online tools
The melting curves can be carried out in all reported software programs for performing qPCR after amplification (Pfaffl MW, 2004). The main objective of the primer is synthesizing DNA with a free terminal end and initiation point of polymerase. A pair of primers one at the template strand while the other at the complementary strand binds on the opposite ends of the sequence being designed, likewise, the 3’ corresponds to the template strand for the process of elongation. The maximum number of candidate primer pairs to screen in order to find specific primer pairs (The candidate primers are generated by primer3 program). Increasing this number can increase the chance of finding a specific primer pair but the process will take longer. The minimal number of contiguous nucleotide base matches between the query sequence and the target sequence that is needed for BLAST to detect the targets.
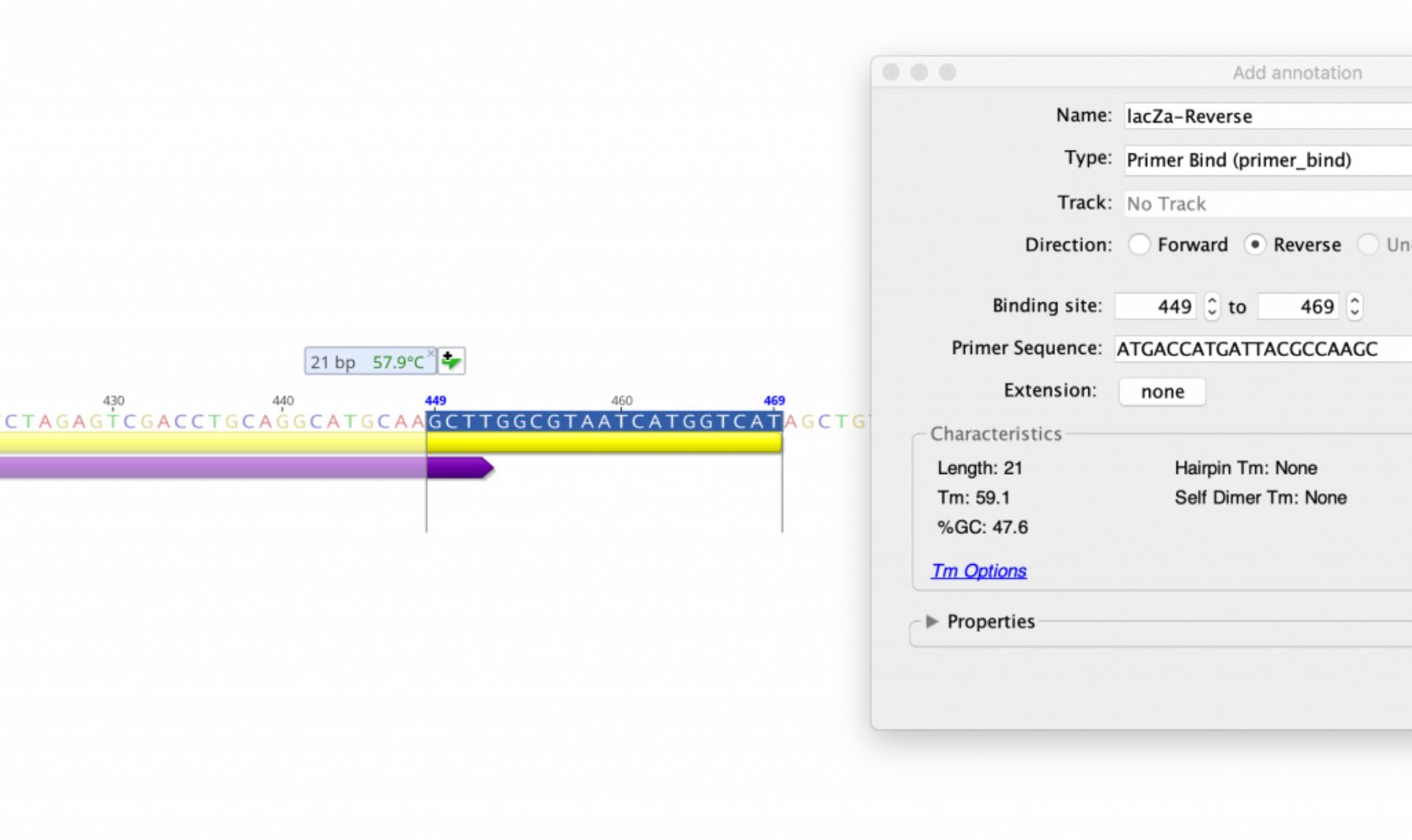
How to: Design PCR primers and check them for specificity
Primer design is a crucial initial step in any experiment utilizing PCR to target and amplify a known nucleotide sequence of interest. Properly designed primers will increase PCR amplification efficiency as well as isolate the targeted sequence of interest with higher specificity. Many factors that may limit the success of a primer pair can be detected a priori with computational methods. For example, primer dimer detection, amplification of alternative products, stem loop interference, extreme melting temperatures, and genotype-specific variations in the target sequence can all be considered computationally to minimize subsequent PCR failures.
Tools
This method demands the use of a vector assembly (plasmid) into a single construct with one or multiple DNA fragments. The PCR primers overlap to form restriction sites with adjacent DNA fragments and are designed, however, Type 2S enzyme, along with DNA ligases of the fragments for a directional assembly. Likewise, this method exploits the use of type 2 class of restriction sites, i.e. cut outside of their restriction sites through non-palindromic sticky end overhangs.
SuperSelective primer pairs for sensitive detection of rare somatic mutations Scientific Reports - Nature.com
SuperSelective primer pairs for sensitive detection of rare somatic mutations Scientific Reports.
Posted: Wed, 17 Nov 2021 08:00:00 GMT [source]
Products and services
Here at NEB, we have created a variety of interactive tools to help you accurately design primers to suit your specific needs. Tm (product)- It measures the melting temperature of the PCR product. Solely living organisms utilize RNA primers while invitro involves DNA primers. However, DNA primers are much preferred due to varied reasons such as stability, easy storage, fewer enzymes required to initiate synthesis. NEB LAMP Primer Design Tool can be used to design primers for your Loop-mediated Isothermal Amplification.

Primer Design and Fragment Assembly Using NEBuilder HiFi DNA Assembly® or Gibson Assembly®
Another example is, when you enter a raw sequence as PCR template rather than an accession, Primer-BLAST will likely report many off-targets. However, some of these might be your intended target that you should choose to allow. When Primer-BLAST does not need to avoid the highly similar off-targets in selecting unique primer regions, it typically has a much higher chance to find specific primers for your intended target.
One also needs to avoid primer-primer annealing which creates primer dimers and disrupts the amplification process. When designing, if unsure about what nucleotide to put at a certain position within the primer, one can include more than one nucleotide at that position termed a mixed site. One can also use a nucleotide-based molecular insert (inosine) instead of a regular nucleotide for broader pairing capabilities. This requires at least one primer (for a given primer pair) to have the specified number of mismatches to unintended targets. The larger the mismatches (especially those toward 3' end) are between primers and the unintended targets, the more specific the primer pair is to your template (i.e., it will be more difficult to anneal to unintended targets). However, specifying a larger mismatch value may make it more difficult to find such specific primers.
This requires that the left or the right primers to span a junction that is just 3' of any such positions. For example, entering "50 100" would mean that the left or the right primers must span the junction between nucleotide position 50 and 51 or the junction between position 100 and 101 (counting from 5' to 3'). You can also specify in the fields below the minimal number of nucleotides that the left or the right primer must have on either side of the junctions.
NEB Primer Design Tools
One must selectively block and unblock repeatedly the reactive groups on a nucleotide when adding a nucleotide one at a time. The main property of primers is that they must correspond to sequences on the template molecule (must be complementary to template strand). However, primers do not need to correspond to the template strand completely; it is essential, however, that the 3’ end of the primer corresponds completely to the template DNA strand so elongation can proceed. Usually a guanine or cytosine is used at the 3’ end, and the 5’ end of the primer usually has stretches of several nucleotides. Also, both of the 3’ ends of the hybridized primers must point toward one another.
Analyzing primer dimer formation is the primary important caution to be taken care of. However, it involves the determination of Gibbs free energy which aids to be the one. Melting temperature (52°C-56°C) The GC results of the sequence gives a fair indication of the primer Tm. Low complexity regions are some regions in a DNA sequence that have biased base compositions such as a stretch of ACACACACACACACACACA. This argument is considered only if Concentration of divalent cations is specified.
Set a lower value if you need to find target sequences with more mismatches to your primers. NEBuilder Assembly Tool can be used to design primers for your Gibson Assembly reaction, based on the entered fragment sequences and the polymerase being used for amplification. NEBuilder Assembly Tool can be used to design primers for your NEBuilder® HiFi DNA Assembly or Gibson® Assembly reaction, based on the entered fragment sequences and the polymerase being used for amplification. The maximum number of PCR targets (amplicons) to be shown when checking specificity for pre-designed primers. The Tm calculation is controlled by Table of thermodynamic parameters and Salt correction formula (under advanced parameters). The default Table of thermodynamic parameters is "SantaLucia 1998" and the default Salt correction formula is "SantaLucia 1998" as recommended by primer3 program.
Primer is a short stretch of sequence that serves as an initiation point for DNA synthesis. There can be a set of primers (forward and reverse) with a sequence complementary to the template DNA -a point of initiation synthesis. With this option on, the program will automatically retrieve the SNP information contained in template (using GenBank accession or GI as template is required) and avoid choosing primers within the SNP regions. You can choose to exclude sequences in the selected database from specificity checking if you are not concerned about these.
This method exploits multiple fragments of DNA by using a combination of overhang sequences on their insert fragments for easy annealing with the adjacent ones. However, type 2S restriction sites permit for golden gate assembly thus leaving no restriction sites after cloning. This controls whether the primer should span an exon junction on your mRNA template. The option "Primer must span an exon-exon junction" will direct the program to return at least one primer (within a given primer pair) that spans an exon-exon junction. You can also exclude such primers if you want to amplify mRNA as well as the corresponding genomic DNA. The structure of the primer should be relatively simple and contain no internal secondary structure to avoid internal folding.
There are critical application-specific parameters to consider that can vastly increase your likelihood of experimental success. Enter the position ranges if you want the primers to be located on the specific sites. The positions refer to the base numbers on the plus strand of your template (i.e., the "From" position should always be smaller than the "To" position for a given primer).
This enables our new graphic display that offers enhanced overview for your template and primers. This specifies the max amplicon size for a PCR target to be detected by Primer-BLAST. In general, the non-specific targets become less of a concern if their sizes are very large since PCR is much less efficient for larger amplicons. The maximum number of Gs or Cs allowed in the last five 3' bases of a left or right primer. The maximum stability for the last five 3' bases of a left or right primer. This specifies the range of total intron length on the corresponding genomic DNA that would separate the forward and revervse primers.
For example, if you want the PCR product to be located between position 100 and position 1000 on the template, you can set forward primer "From" to 100 and reverse primer "To" to 1000 (but leave the forward primer "To" and reverse primer "From" empty). Note that the position range of forward primer may not overlap with that of reverse primer. One needs to design primers that are complementary to the template region of DNA.

No comments:
Post a Comment